What is DendroTweaks?
DendroTweaks is a Python-based toolbox designed to streamline the creation and validation of single-cell biophysical neuronal models. It is available as both a standalone Python package and a web-based application, offering flexibility for diverse user needs.
| PyPI Package | https://pypi.org/project/dendrotweaks/ |
| Documentation | https://dendrotweaks.readthedocs.io/en/latest/index.html |
| GitHub Repository (Package) | https://github.com/Poirazi-Lab/DendroTweaks |
| GitHub Repository (Web Application) | https://github.com/Poirazi-Lab/DendroTweaksApp |
To learn more about the toolbox, you can read our publication in eLife.
Roman Makarov, Spyridon Chavlis, Panayiota Poirazi (2024). DendroTweaks: An interactive approach for unraveling dendritic dynamics. eLife, 13:RP103324.
If you find DendroTweaks helpful for building your models, please consider citing our work.
@article{Makarov2024,
title={DendroTweaks: An interactive approach for unraveling dendritic dynamics},
author={Makarov, Roman and Chavlis, Spyridon and Poirazi, Panayiota},
journal={eLife},
volume={13},
pages={RP103324},
year={2024},
doi={10.7554/eLife.103324.1}
}For a quick overview of the toolbox, including a video demonstration, you can visit our e-poster presented at the FENS Forum 2024 in Vienna.
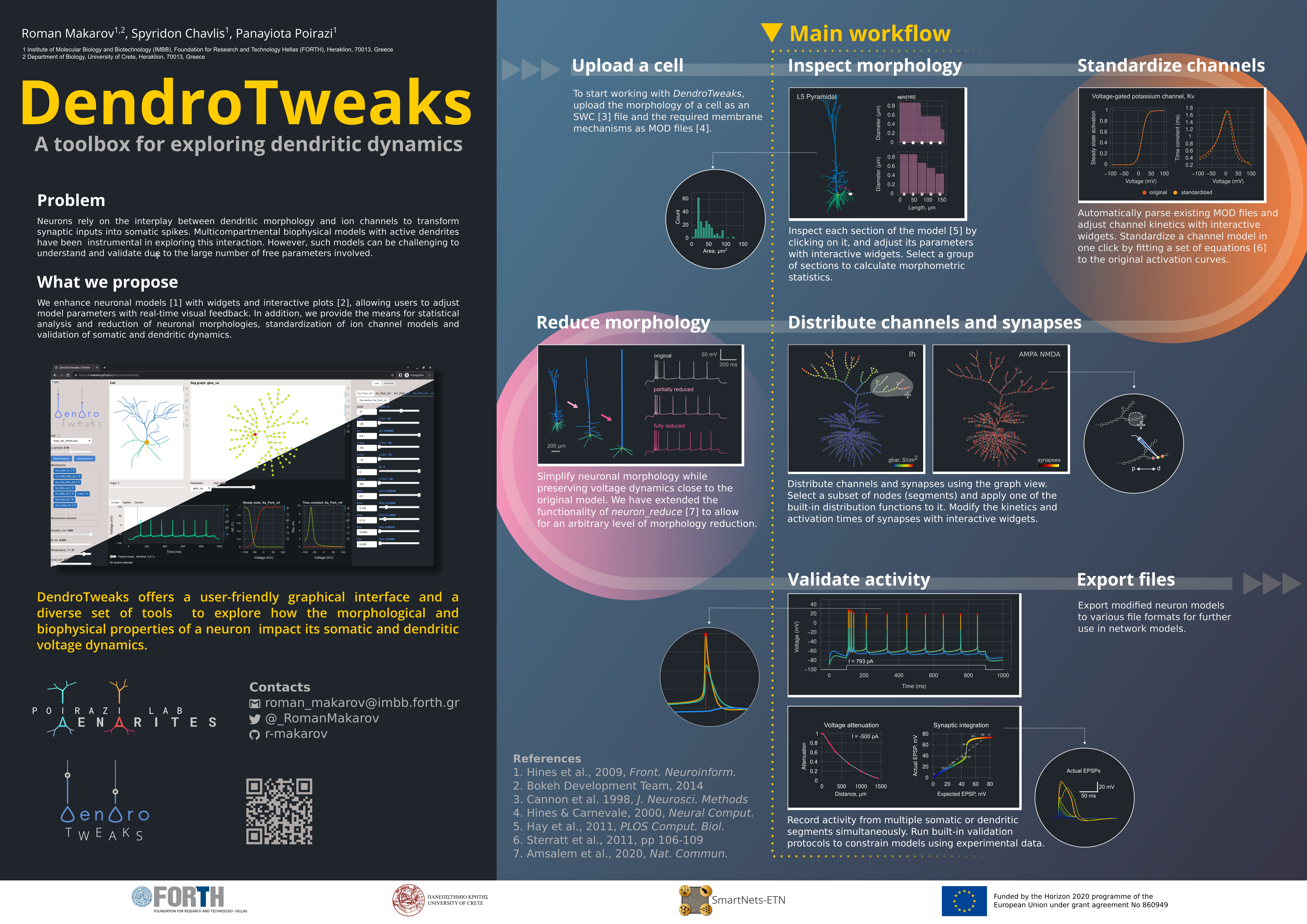
Key Features
Visualization
DendroTweaks offers an intuitive interface for visualizing and analyzing single-neuron models. You can visualize the morphology, ion channel distributions, and synaptic inputs as well as the resulting activity of the neuron.
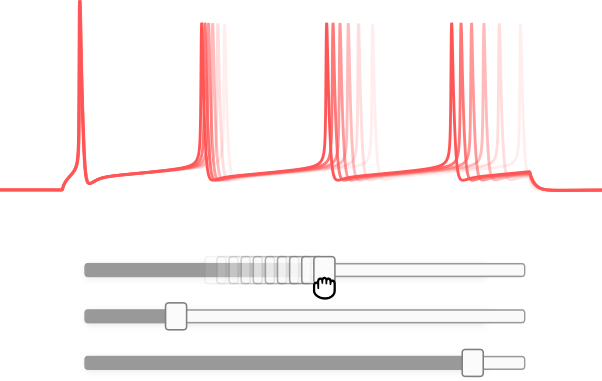
Interactive simulations
DendroTweaks allows you to adjust any parameter of the model and receive real-time visual feedback. With widgets and interactive plots, you can explore the impact of different morphological and biophysical parameters on the activity of a neuron.
Standardization of ion channel models
You can automatically convert existing
.mod files into Python classes.
Additionally, the toolbox allows you to adjust channel kinetics
with interactive widgets and standardize ion channel models by fitting
a set of equations [1] to the original kinetic profiles.
Morphology reduction
You can reduce neuronal morphology by simplifying the
dendritic structure. We have extended the functionality
of neuron_reduce
[2] to allow for an arbitrary
level of morphology reduction.
Automated validation
Our toolbox provides a set of built-in validation protocols to ensure the model's activity is consistent with the experimental data.
- Principles of Computational Modelling in Neuroscience (Sterratt, D. et al.; 2011)
- Amsalem, O., Eyal, G., Rogozinski, N. et al. An efficient analytical reduction of detailed nonlinear neuron models. Nat Commun 11, 288 (2020). https://doi.org/10.1038/s41467-019-13932-6
Contacts
DendroTweaks was created by Roman Makarov and is currently maintained by Roman Makarov and Spyridon Chavlis, both members of the Poirazi Lab.
For any questions or suggestions, please feel free to contact us at: dendrotweaks@dendrites.gr
